Hi Will et al.! I ran MaAsLin 3 on 21k transcripts across 561 metatranscriptomes, in models with fixed and random effects:
maaslin3(input_data = counts,
input_metadata = metadata,
feature_specific_covariate = magabunds,
feature_specific_covariate_record = FALSE,
feature_specific_covariate_name = "magabund",
min_prevalence = 0.19,
output = output,
normalization = 'TSS',
transform = 'LOG',
standardize = TRUE,
plot_summary_plot = FALSE,
plot_associations = FALSE,
formula = '~ arm + study.wk + arm:study.wk + sex + cohort + age_enroll + mtxseqdepth + (1|pid)')
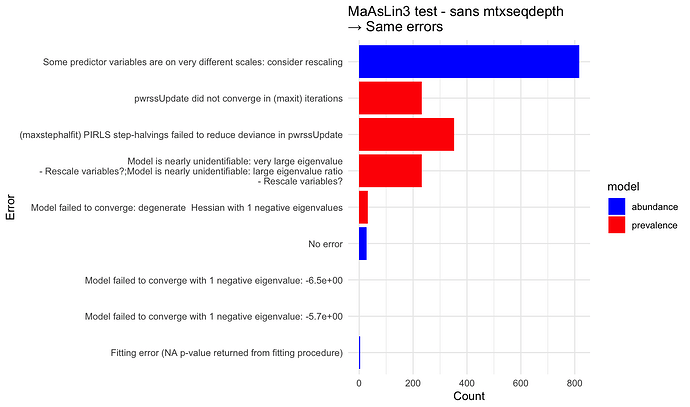
Unfortunately the majority of the models yielded errors, summarized here:
Higher-prevalence transcripts were more likely to have error-free models, though there were many exceptions (i.e., transcripts close to my 0.19 prevalence cutoff whose models nonetheless did not always error):
My team and I would be thrilled to use this tool for this current and future projects if these errors could be troubleshot:
Some predictor variables are on very different scales: consider rescaling
→ I thought standardize == TRUE is supposed to fix this? I tried pre-standardizing the data withmaaslin_process_metadata; that did not fix it.(maxstephalfit) PIRLS step-halvings failed to reduce deviance in pwrssUpdate
→ Does the link function need to be changed?pwrssUpdate did not converge in (maxit) iterations
→ If I understand correctly that this is due to zero inflation, I thought maaslin3 was built to handle that?Model is nearly unidentifiable: very large eigenvalue\n - Rescale variables?;Model is nearly unidentifiable: large eigenvalue ratio\n - Rescale variables?
→ See the above comment about standardize == TRUEFitting error (NA p-value returned from fitting procedure)Model failed to converge with 1 negative eigenvalue: XModel failed to converge with max|grad| = X (tol = Y, component 1)
→ These last 3 errors are less prevalent, but they’d still be great to address
Thanks so much in advance!
Best regards,
Liam Fitzstevens
Gordon Lab, WashU Medical School in St. Louis
Hi Liam,
Thanks for asking about this!
A few thoughts and questions:
- I don’t know how many arms you have and what the number of covariates is in the model matrix (i.e., categorical variables * categories per variable), but just by reading the formula, it looks like this might be a very complicated model. We’ve found in our own runs that the mixed effects logistic models with complicated formulas often produce errors just because there are so many parameters the model is trying to optimize. Usually, this is due to using random effects with only a small number of samples per subject: I would wager that running the model without
(1|pid) gets rid of a lot of the errors. To then control for pid, there are a few options.
- Simply including
pid as a fixed effect would certainly control for pid, but it might make some of your effects lose significance. It’s worth trying including pid as a covariate, removing all the pid results from the outputs, and BH correcting the p-values for the fixed coefficients that actually are of interest. However, if the coefficients you care about never (or rarely) change within a pid, this won’t work.
- If most individuals have a single measurement, it might be worth averaging or subselecting one measurement per individual and running a model without
pid.
- If neither of these is the case, let’s fix the scale issue and come back to it.
- Different scale: When you tried pre-standardizing with
maaslin_process_metadata, did the continuous variables come out properly standardized (i.e., mean 0 variance 1)? If not, it’s a bug on our end. If they’re fine, can you run the model without mtxseqdepth to see if mtxseqdepth is the source of the issue (because it has the widest range)? It’s also possible that scaling is actually causing problems if, for example, one of your continuous variables has values that are almost always the same but occasionally different, yielding standardized values with many near-zeros and a few extreme z-scores.
PIRLS step-halvings: This is probably a symptom of one of the above rather than its own issue. Under the hood, if model fitting fails, MaAsLin 3 calls a different logistic regression optimizer until it runs out of optimizers to try. It also uses a maximum number of iterations that’s much larger than the default, so I doubt that changing the optimizer or iterations will help much. The link function is inherent to logistic regression or linear regression, so that should stay.pwrssUpdate...: If the issue was due to uncontrolled zero-inflation, the model would fit but with very extreme coefficients and standard errors (since they’re shooting off to infinity because of linear separability). I think this is also a symptom of one of the first two issues rather than the zero inflation.Model if nearly unidentifiable... is probably the scaling issue.Fitting error (NA... usually means that the model “fit correctly” but that something was wrong with the model specification (e.g., there was perfect collinearity in the model matrix). Likely the only way to fix this is to change the formula.Model failed to converge... is probably caused by the same issues as above.
Let me know what you find on those first two points. I’m happy to troubleshoot more or maybe look at a reproducible snippet of the data if it’s still not working!
Will
Hi Will,
Thank you so much for such a comprehensive answer!
- Manually running
maaslin_process_metadata produced mean 0s & variance 1s, indicating no bug on your end:

If they’re fine, can you run the model without mtxseqdepth to see if mtxseqdepth is the source of the issue
It’s also possible that scaling is actually causing problems if, for example, one of your continuous variables has values that are almost always the same but occasionally different, yielding standardized values with many near-zeros and a few extreme z-scores.
As an experiment, I tried raw metadata (i.e., not processed by maaslin_process_metadata):
Removing feature_specific_covariate (DNA abundances) ended up doing the trick!
I had assumed up to this point that transform = 'LOG' also applied to feature_specific_covariate, but realized this may not be the case. Manually log-transforming the feature_specific_covariate dataframe fixed it!
You were right about 3. - 7. being symptoms of the actual problem - this is fantastic. Thanks again, Will!
All the best from STL,
Liam
Ha - what a red herring!
The preprocess_dna_mtx function (mentioned here) is supposed to take care of the upstream transformations. If you didn’t use that function, it might be worth re-running with the transformations from that function since it also makes sure to properly handle cases in which one or both of the DNA and RNA are 0.
Will
Ah, I thought preprocess_dna_mtx was baked into maaslin3 - thanks for pointing that out! Done:
NA p-value returned from fitting procedure seems entirely normal. Thanks again!!
1 Like
I realize that preprocess_dna_mtx cannot be run in cases like mine, where one uses genome-level (species) DNA abundances as covariates instead of gene-level DNA abundances (as was the case in your HMP example). This is because in my case, multiple genes belong to the same genome, upon which a TSS (i.e., the first step of preprocess_dna_mtx) cannot be performed.
I am happy to perform TSS at the genome-level manually and perform preprocess_dna_mtx’s remaining 3 steps myself:
With that said, I imagine that many users will also find themselves in my position; with DNA abundances at the genome- rather than gene-level. If you agree, it might be nice to break preprocess_dna_mtx down into something like preprocess_genelvldna_mtx and preprocess_genomelvldna_mtx. The latter would require an additional column in dna_table that identifies the genome that each transcript belongs to, and to perform the TSS on the genomes’ abundances per participant.
What do you think? I only bring this up as I think it might help catalyze field-wide adoption.
Hi Liam,
Thanks for the suggestion! Are you saying that your RNA table is species-gene by sample and your DNA table is species by sample? Out of curiosity, what did you use upstream to get those tables? The reason we’ve implemented it this way is that we assumed most people would have used HUMAnN for both DNA and RNA, which will give species-gene by sample tables.
Will
Hi Will,
Yes, that’s correct. The DNA and RNA counts were both calculated using Kallisto and a set of species representative genomes (SRGs; a 95% ANI-dereplicated MAG set).
The scenario I described would also apply to people doing metagenomic profiling of SRGs with Kraken/Bracken, of which I think there are many.
If you don’t implement this modification, future users falling under this use case can perform the following work-around:
- Reformat the DNA table from gene- back to MAG-level.
- Run Maaslin3’s
TSSnorm function on the MAG abundances manually, rather than calling it as a part of preprocess_dna_mtx as is standard.
- Reformat the output of Step 2 to again be by gene.
- Remove the code in
preprocess_dna_mtx that TSSnormalizes DNA.
- Plug the output of Step 3 and the RNA counts into the modified
preprocess_dna_mtx function from Step 4.
Liam
Hi Liam,
To handle this case, I’ve added the function preprocess_taxa_mtx which works similarly to preprocess_dna_mtx but applies to the taxon level rather than the gene level. It takes a table of taxonomic abundances, a table of per-gene-per-taxon RNA abundances, and a table with the name of the corresponding taxon for each RNA. It performs the same steps as preprocess_dna_mtx except when the taxon abundance is zero and any of that taxon’s RNA abundances are non-zero, it imputes half the dataset minimum for that taxon. This yields model 5 from the lab’s MTX paper, which typically performed as well as preprocess_mgx_mtx (model 6).
The documentation is available with ?preprocess_taxa_mtx, and a note is added on the MaAsLin 3 MTX model wiki. This is currently available with the GitHub installation and should be available from the Bioconductor installation soon.
Will
Hi Will,
Fantastic! I imagine this will help with adoption as I don’t think this is an atypical use-case. Thanks again for your help with the errors!
Cheers,
Liam